Practical reporting guidelines, summary of key concepts, test selection parameters table, multiple comparison corrections table, and scipy.stats functions reference. Complete reference guide for hypothesis testing.

This article is part of the free-to-read Machine Learning from Scratch
Choose your expertise level to adjust how many terms are explained. Beginners see more tooltips, experts see fewer to maintain reading flow. Hover over underlined terms for instant definitions.
Summary and Practical Guide to Hypothesis Testing
In 1925, Ronald Fisher published Statistical Methods for Research Workers, introducing hypothesis testing to the scientific world. His framework revolutionized how we learn from data, providing a rigorous method for distinguishing signal from noise. Nearly a century later, hypothesis testing remains the backbone of empirical research across every scientific discipline: from medicine and psychology to economics and machine learning.
Yet despite its ubiquity, hypothesis testing is frequently misused and misunderstood. Studies show that many published papers contain statistical errors, misinterpret p-values, or fail to report essential information. The goal of this final chapter is to synthesize everything you've learned into a practical guide that helps you avoid these pitfalls and conduct hypothesis tests that are both valid and useful.
This chapter serves as your reference manual: a complete framework for choosing the right test, conducting the analysis correctly, and reporting results in a way that advances scientific knowledge. Keep it handy whenever you're working with data.
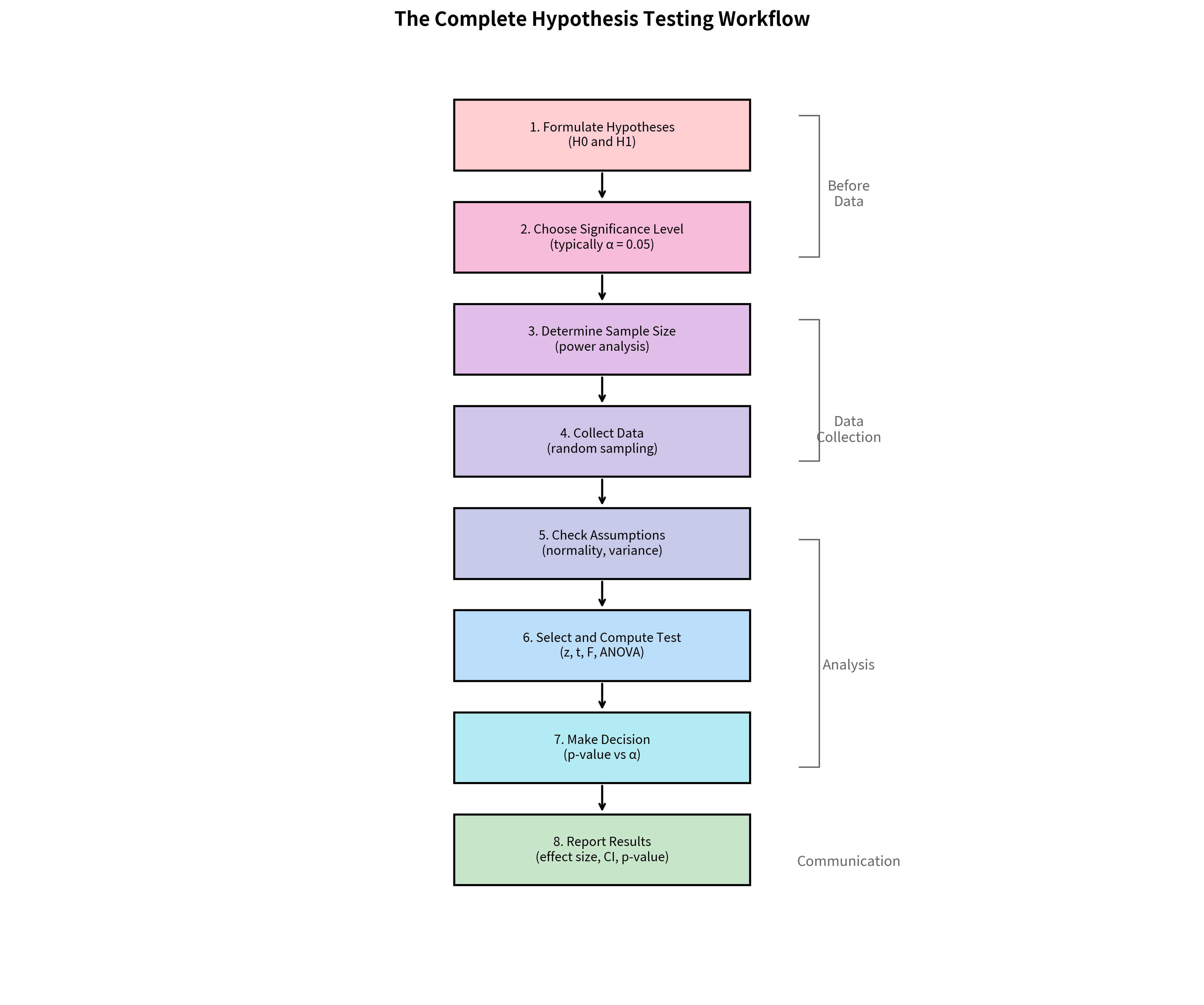
The Complete Hypothesis Testing Workflow
Before diving into details, here's the complete workflow for conducting a hypothesis test. Each step is critical: skipping any one can invalidate your conclusions.
Step-by-Step Guide
Step 1: Formulate Hypotheses
- Define (null hypothesis): The default assumption, typically "no effect" or "no difference"
- Define (alternative hypothesis): What you're trying to demonstrate
- Choose one-tailed or two-tailed based on your research question
- Do this BEFORE seeing the data
Step 2: Choose Significance Level (α)
- Standard: α = 0.05 (5% false positive rate)
- Stringent: α = 0.01 for high-stakes decisions
- Lenient: α = 0.10 for exploratory research
- Consider the consequences of Type I vs Type II errors
Step 3: Determine Sample Size
- Use power analysis to calculate required n
- Specify: α, power (typically 0.80), minimum effect size of interest
- Balance statistical needs against practical constraints
Step 4: Collect Data
- Use appropriate randomization and sampling methods
- Ensure independence of observations
- Avoid peeking at results during collection
Step 5: Check Assumptions
- Normality: Shapiro-Wilk test, Q-Q plots
- Equal variances: Levene's test
- Independence: Study design consideration
- Choose robust alternatives if assumptions violated
Step 6: Select and Compute Test
- Use the decision tree in the next section
- Calculate test statistic and p-value
- Compute confidence interval
Step 7: Make Decision
- If p < α: Reject H₀, conclude evidence for H₁
- If p ≥ α: Fail to reject H₀, insufficient evidence
- Remember: "fail to reject" ≠ "accept H₀"
Step 8: Report Results
- Effect size (Cohen's d, η², r)
- Confidence interval
- Exact p-value
- Test statistic and degrees of freedom
- Assumption check results
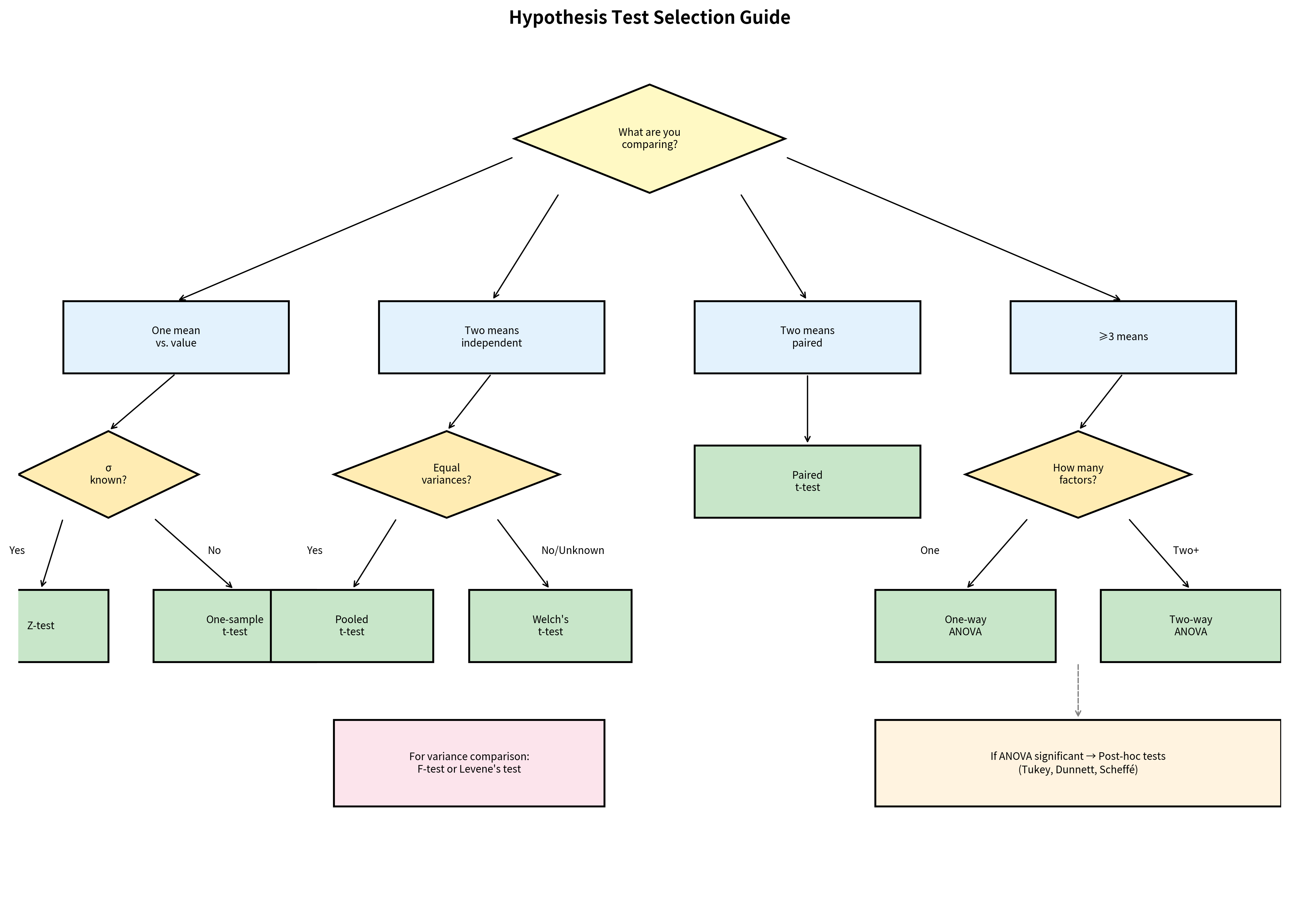
Test Selection Decision Tree
Choosing the correct test is critical. Use this decision framework based on your research question and data characteristics.
Quick Reference Table
| Research Question | Test | scipy.stats Function |
|---|---|---|
| Is the mean equal to a specific value? (σ known) | Z-test | Manual calculation |
| Is the mean equal to a specific value? (σ unknown) | One-sample t-test | ttest_1samp() |
| Are two independent group means equal? | Welch's t-test | ttest_ind(equal_var=False) |
| Are two independent group means equal? (equal var) | Pooled t-test | ttest_ind(equal_var=True) |
| Are paired/matched observations different? | Paired t-test | ttest_rel() |
| Are ≥3 group means equal? | One-way ANOVA | f_oneway() |
| Are two variances equal? | F-test | f.sf() (manual) |
| Are multiple variances equal? | Levene's test | levene() |
| Which pairs differ after ANOVA? | Tukey HSD | tukey_hsd() |
| Treatment vs control comparisons? | Dunnett's test | dunnett() |
Effect Size Reference
Effect sizes quantify the magnitude of an effect independent of sample size. Always report them alongside p-values.
Cohen's d (Comparing Two Means)
| Cohen's d | Interpretation | Practical Example |
|---|---|---|
| 0.2 | Small | Subtle difference, requires large sample to detect |
| 0.5 | Medium | Noticeable effect, visible with moderate sample |
| 0.8 | Large | Substantial effect, obvious in most analyses |
| 1.2+ | Very large | Major effect, visible to naked eye |
Related measures:
- Hedges' g: Corrects d for small sample bias
- Glass's Δ: Uses control group SD only
- Cohen's d_z: For paired designs, uses SD of differences
ANOVA Effect Sizes
| Measure | Formula | Interpretation |
|---|---|---|
| η² (eta-squared) | % variance explained (biased upward) | |
| ω² (omega-squared) | Corrected formula | Less biased estimate |
| Partial η² | For factorial designs | Effect controlling for other factors |
Benchmarks for η² and ω²:
- Small: 0.01
- Medium: 0.06
- Large: 0.14
Correlation as Effect Size
| r | Interpretation | r² (variance explained) |
|---|---|---|
| 0.1 | Small | 1% |
| 0.3 | Medium | 9% |
| 0.5 | Large | 25% |
Power Analysis Quick Reference
Power analysis determines the sample size needed to detect an effect of a given size with specified probability.
The Power Pentagon
Five quantities are interconnected: knowing any four determines the fifth:
- Sample size (n): Number of observations
- Effect size (d): Magnitude of the effect
- Significance level (α): False positive rate
- Power (1-β): True positive rate
- Variability (σ): Data spread
Sample Size Formulas
One-sample t-test:
Two-sample t-test (equal groups):
Two proportions:
Sample Size Table (Two-Sample t-test, α = 0.05, Two-tailed)
| Effect Size | Power = 0.80 | Power = 0.90 | Power = 0.95 |
|---|---|---|---|
| d = 0.2 (small) | 394 per group | 527 per group | 651 per group |
| d = 0.5 (medium) | 64 per group | 86 per group | 105 per group |
| d = 0.8 (large) | 26 per group | 34 per group | 42 per group |
Multiple Comparisons Reference
When to Use Each Method
| Method | Controls | Use When |
|---|---|---|
| Bonferroni | FWER | Few tests, any false positive costly |
| Holm | FWER | Many tests, need more power than Bonferroni |
| Benjamini-Hochberg | FDR | Exploratory analysis, many tests OK |
| Tukey HSD | FWER | All pairwise comparisons after ANOVA |
| Dunnett | FWER | Comparing treatments to control |
Quick Formulas
Bonferroni: Reject if
Holm (step-down): For ordered p-values, reject if
Benjamini-Hochberg: Find largest where , reject all
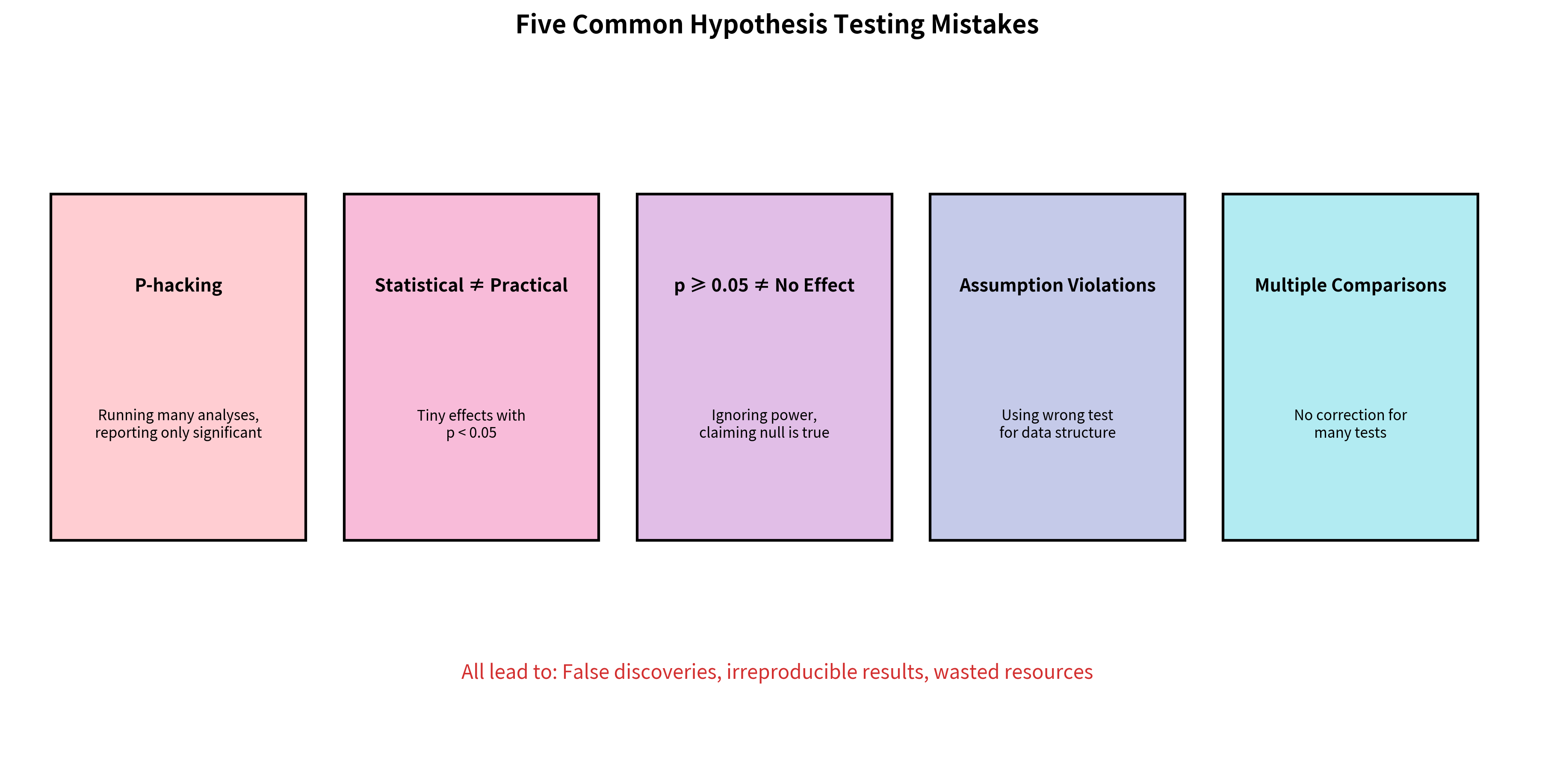
Common Mistakes and How to Avoid Them
Mistake 1: P-Hacking
Problem: Running multiple analyses and only reporting significant results.
Solution: Pre-register your analysis plan. Report all tests conducted, not just significant ones. Use appropriate multiple comparison corrections.
Mistake 2: Confusing Statistical and Practical Significance
Problem: Treating p < 0.05 as proof that an effect matters.
Solution: Always report effect sizes. Ask "Is this effect large enough to be meaningful?" even when statistically significant.
Mistake 3: Misinterpreting Non-Significant Results
Problem: Concluding "no effect exists" when p ≥ 0.05.
Solution: Consider statistical power. Report confidence intervals to show the range of plausible effects. Distinguish "evidence of absence" from "absence of evidence."
Mistake 4: Violating Assumptions
Problem: Using parametric tests when assumptions are violated.
Solution: Check assumptions before testing. Use robust alternatives (Welch's t-test, non-parametric tests) when assumptions fail.
Mistake 5: Ignoring Multiple Comparisons
Problem: Running many tests without correction, inflating false positive rate.
Solution: Plan your analyses in advance. Apply appropriate corrections. Report the number of tests conducted.
Complete Reporting Example
Here's an example of a complete analysis with proper reporting:
scipy.stats Quick Reference
Testing Functions
Distribution Functions
Summary: Key Takeaways
The Foundations
- P-values measure evidence against H₀, not the probability H₀ is true
- Confidence intervals show the range of plausible parameter values
- Effect sizes quantify magnitude independent of sample size
- Power determines your ability to detect effects that exist
The Tests
- Z-test: When σ is known (rare in practice)
- t-test: The workhorse for comparing means
- Welch's t-test: Default for two-group comparisons
- ANOVA: For comparing ≥3 groups
- Post-hoc tests: After significant ANOVA
The Errors
- Type I (α): False positive: rejecting true H₀
- Type II (β): False negative: failing to reject false H₀
- Multiple comparisons: Inflate error rates without correction
The Practice
- Plan before collecting data: Hypotheses, α, sample size
- Check assumptions: Normality, equal variances, independence
- Report completely: Effect size, CI, exact p-value, test used
- Interpret cautiously: Statistical ≠ practical significance
Conclusion
Hypothesis testing is a powerful framework for learning from data, but its power comes with responsibility. The methods you've learned in this series, from basic p-values and confidence intervals to power analysis, effect sizes, and multiple comparison corrections, form a complete toolkit for rigorous statistical inference.
Remember these principles:
- Design before analysis: Plan your hypotheses, tests, and sample size before seeing data
- Check your assumptions: Use appropriate tests for your data structure
- Report completely: Enable others to evaluate and replicate your work
- Think beyond p-values: Effect sizes and confidence intervals tell a richer story
- Control multiplicity: Correct for multiple tests when applicable
Statistics is not about proving things with certainty: it's about quantifying uncertainty and making informed decisions despite incomplete information. Used well, hypothesis testing helps us separate signal from noise and build cumulative scientific knowledge. Used poorly, it generates false discoveries and wastes resources.
The difference between the two lies in understanding not just how to calculate test statistics, but why each step matters and what can go wrong. With the knowledge from this series, you're equipped to conduct hypothesis tests that are valid, interpretable, and useful.
This concludes the hypothesis testing series. For hands-on practice, try applying these methods to your own data, starting with clear hypotheses and working through each step of the workflow.
Quiz
Ready to test your understanding? Take this comprehensive quiz to reinforce what you've learned throughout the hypothesis testing series.



Comments